4. Next Generation Synthesis
Part A - Primer Design
Ok so we need to design two sets of primers. One set will amplify the first part of the amilCP, a choromoprotein. So we open up mUAV which contains this protein on Benchling, and annotate the protein gene and the special area (6 bases) which we can mutate to create different colors.
Lets start with the “first” part of the gene, up to the mutatagensis area. I marked 24 bases, starting from the RBS (Benchling shows you the Melting temp, and only on 24 it was above 48C), I couldn’t underdtand exactly where is the promoter? Then right click and Create Primer (forward).
Next, I went to pUC1, and marked the last 20 bases before PuvII cut site. Copied those over, went back to the primer and pasted them to the beginning of the primer. You could see how 24 fit perfectly while the overhang does not

The length of the primer is 44, the GC content is 34.1%, and the Melting Temp is 61.1C
Next, the reverse primer for the same part, we go to 2309 and mark 32 bases (32 because then I get the same temp of 61.3)

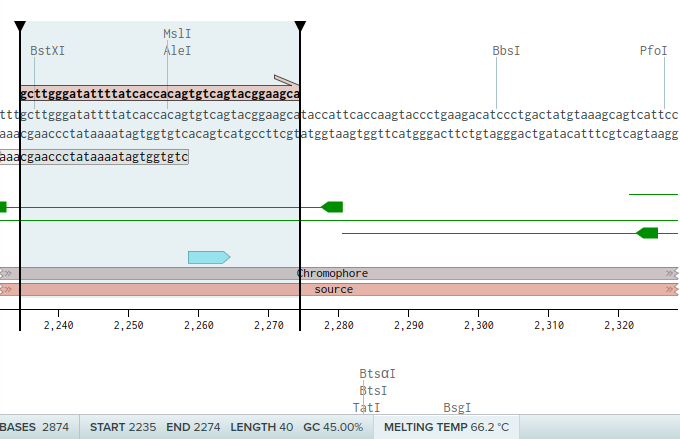
Now for the second set of primers. We want a forward primers which include the mutation site (chromophore), according to instructions we should take 24 bases before, and I decided to take 10 bases after. Resulting in a 40 base long primer, with GC content of 45% and Metling temp of 66C
And for the reverse primer, I selected an area after the terminators that had similar melting temp - 30 bases. Then I went back to pUC1, took the part just where PuvII cuts, and copied the reverse complement. Then I pasted it into primer… but - I got a very high melting temp! 73.1C. Damn.
Second attempt
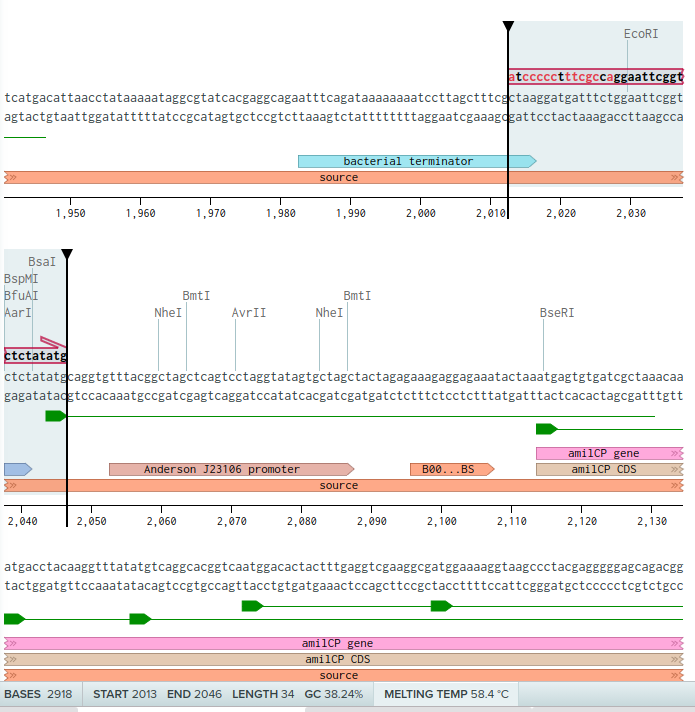
Ok apparently there was some confusion with annotations. So I re-did the whole process described above, but instead of downloading the Genbank file off Addgene, I found the mUAV sequence NCBI GenBank and downloaded it from there (Send to > File). Importing this file into Benchling resulted in a more complete anottation (including the promoter of amilCP).
Again, The first primer is a forward one containing bases just before the promoter
Then it’s reverse primer starts just before the chromophore:
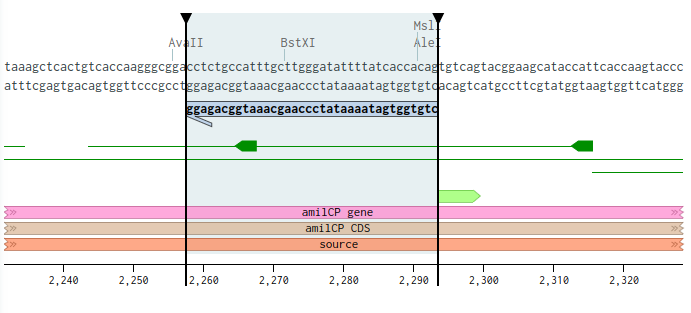
Then for the second set, the forward primer start 24 bases before the chromophore, and 10 bases after

For the last primer, I still had a problem with a very high CG concentration after the third terminator, so I decided to include a small part of the terminator which has lots of Ts. And included 20 bases from the pUC19 backbone:

Overall:

PartA_fwd: ATCCCCCTTTCGCCAGGAATTCGGTCTCTATATG (Tm: 65.1C)
PartA_rev: CTGTGGTGATAAAATATCCCAAGCAAATGGCAGAGG (Tm: 63.8C)
PartB_fwd_orig: GCTTGGGATATTTTATCACCACAGTGTCAGTACGGAAGCA (Tm: 66.8C)
PartB_fwd_mut1: GCTTGGGATATTTTATCACCACAGGTCAACTACGGAAGCA (Tm: 64.9C)
PartB_fwd_mut2: GCTTGGGATATTTTATCACCACAGTTTATGTACGGAAGCA (Tm: 62.9C)
...
PartB_fwd_mut7: GCTTGGGATATTTTATCACCACAGTCAATGTACGGAAGCA (Tm: 64C)
PartB_rev: CCTCTTCGCTATTACGCCAGATAACGCGAAGTAATCTTTT (Tm: 65.2C)
(Melting temperatures were derived using Benchling as well as IDT Oligo order form)
Part B - Experiment
Protocol
1) Follow the QIAprep Spin Miniprep Kit protocol, using 3ml of inital cell culture to start with.
After nanodrop, I got 99.7 ng/ul concentration of mUAV DNA.

2) Digest the pUC19 plasmid with the restriction enzyme PvuII to generate the linear blunt-ended backbone fragment. Prepare the reaction in a PCR tube (add enzyme last and water first):
| To Reaction | Volume (50 uL Totral) |
| DNase/RNase-Free Water | 40 uL |
| 2 ug pUC19 (1 ug/ul in green cap tubes) | 2 uL |
| 10x NEB Buffer CutSmart | 5 uL |
| NEB PvuII-HF | 3 uL |
Incubate reactions at 37C for at least 15 minutes in thermocycler. and put in the fridge.
3) Amplify PCR fragments for the gene assembly of amilCP mutants.
Prepare the reaction in a PCR tube to produce the universal chromophore-preceding fragment:
| To Reaction | Volume (50 uL Total) |
| DNase/RNase-Free Water | 18 uL |
| 200 ng miniprepped amilCP | 2 uL |
| Forward primer with tail that overlaps with cut pUC19 (10 uM in yellow cap labeled “TailVF Fwd”) | 2.5 uL |
| Reverse primer preceding the chromophore (10 uM in yellow cap labeled “CPUni Rev”) | 2.5 uL |
| 2x Phusion High-Fidelity PCR Master Mix | 25 uL |
Prepare the reaction in a PCR tube to produce the chromophore mutant library fragments:
| To Reaction Volume | (50 uL Total) |
| DNase/RNase-Free Water | 18 uL |
| 200 ng miniprepped amilCP | 2 uL |
| Reverse primer with tail that overlaps with cut pUC19 (10 uM in purple cap labeled “TailVR Rev”) | 2.5 uL |
| Forward primer defining the chromophore library and overlaps with the universal reverse primer (10 uM in purple cap labeled “CPLib Fwd”) | 2.5 uL |
| 2x Phusion High-Fidelity PCR Master Mix | 25 uL |
Thermocycle reactions in a PCR program with with following program.
- Initial heat denature DNA at 98C for 30 seconds
- Repeat the following for 35 cycles:
- Denature DNA at 98C for 10 seconds
- Anneal primers at 60C for 20 seconds
- Extend from primers with DNA polymerase primers at 72C for 1 minute. (Account for 1kb/30sec processivity)
- Final extension at 72C for 5 minutes.
- Hold at 4C until samples are retrieved.

4) Purify the DNA products from your reactions using the Zymo DNA Clean & Concentrator™-25 Kit. The following protocol is copied from Zymo Research and based on silica adsorption.
- For each reaction: In a 1.5 ml microcentrifuge tube, add 5x volumes (250ul) of DNA Binding Buffer to each volume of reaction. Mix briefly by vortexing.
- Transfer each mixture into a separate Zymo-Spin™ Column in a Collection Tube.
- Centrifuge for 30 seconds at 13,000 rpm (~17,900 x g). Discard the flow-through.
- Add 200 µl DNA Wash Buffer to the column. Centrifuge at for 30 seconds at 13,000 rpm (~17,900 x g). Repeat the wash step.
- Transfer the column to a new 1.5 ml microcentrifuge tube and add 25 µl of DNase/RNase-free water directly to the column matrix. Let sit at room temperature for one minute.
- Centrifuge for 30 seconds at 13,000 rpm (~17,900 x g) to elute.
5) Measure the concentration of your purified DNA using the Nanodrop spectrophotometer.
I got the following concentrations:
- CPUni (universal part of the chromoprotein): 8.6 ng/uL
- CPLib (mutated part with the chromophore): 6.3 ng/uL
6) Setup your Gibson Assembly reaction in PCR tubes.
- In a
10 ultotal volume, mix 100 ng of cut vector (PvuII digest) with a 2-fold excess of gene fragment (PCR reactions) in DNase/RNase-free water. Molar ratios can be approximated by the lengths of the DNA products. Note, the large fragment of the digested pUC19 is 2,364 bp. The gene fragment amplicons are 353 bp and 777 bp.
For the PvuII cut vector, we have 50uL which has 2ug of DNA. So to get 100ng we need get take 2.5uL
The formula to calculate inserts:
For CPUni:
And CPLib:
And then divide by concentrations to get the volume:
- CPUni - 3.5uL
- CPLib - 10.4uL
So if we mix all of it together we get 16.4uL
- Combine with
10 ul16.4 uL 2x Gibson Master Mix. - Incubate in the thermocycler at 50C for at least 15 minutes.
Random action shot:

7) Electroporate assemblies into electrocompetent E coli (stored across the hall in the -80C).
- Move Gibson assembly into a cold block on ice. Keep electroporation cuvettes, 10% Glycerol and microcentrifuge tubes (you can also use a PCR strip) on ice.
- Thaw electrocompetent cells on ice for 15-20 minutes.
- Aliquot 75 ul of 10% Glycerol followed by 25ul of competent cells into microcentrifuge tubes kept in a cold block on ice.
- Add 5 µl of the assembled product to the competent cells. Mix gently by pipetting up and down or by flicking the tube 4–5 times. Do not vortex.
- Transfer the mix to electroporation cuvettes without bubbles. Tap them gently to make sure the mix settles at the bottom part of the cuvette.
- Electroporate at 1700-2000V with 5ms time constant.
- Immediately transfer 900ul of
SOCSOB into the cuvettes. Pipette back and forth. - Transfer it into a new 1.5 ml microcentrifuge tube. Incubate them for one hour at 37C.
- Spread 100-250 µl of the cells onto the ampicillin selection plates.
- Incubate overnight at 37C in the warm room.
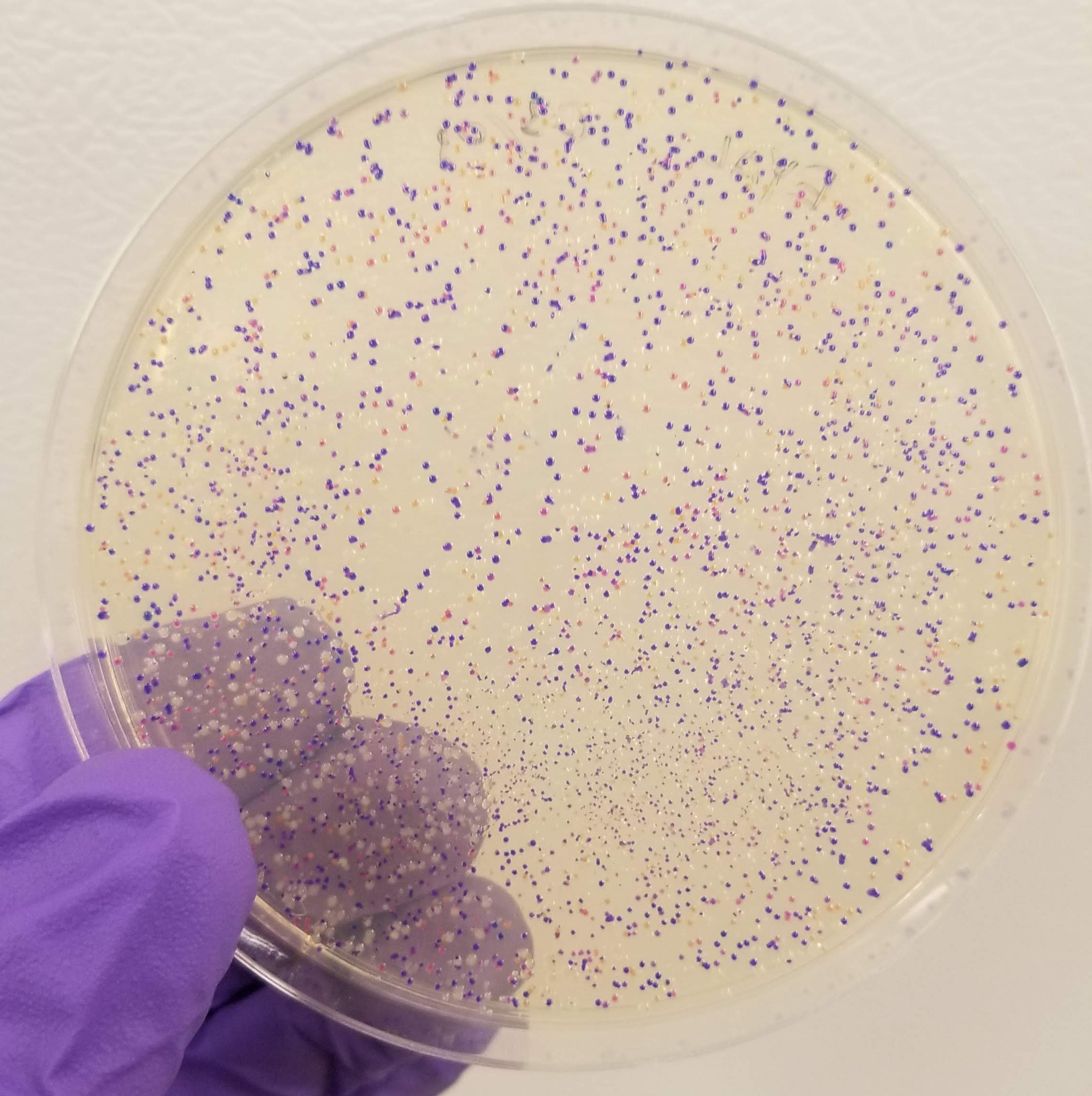
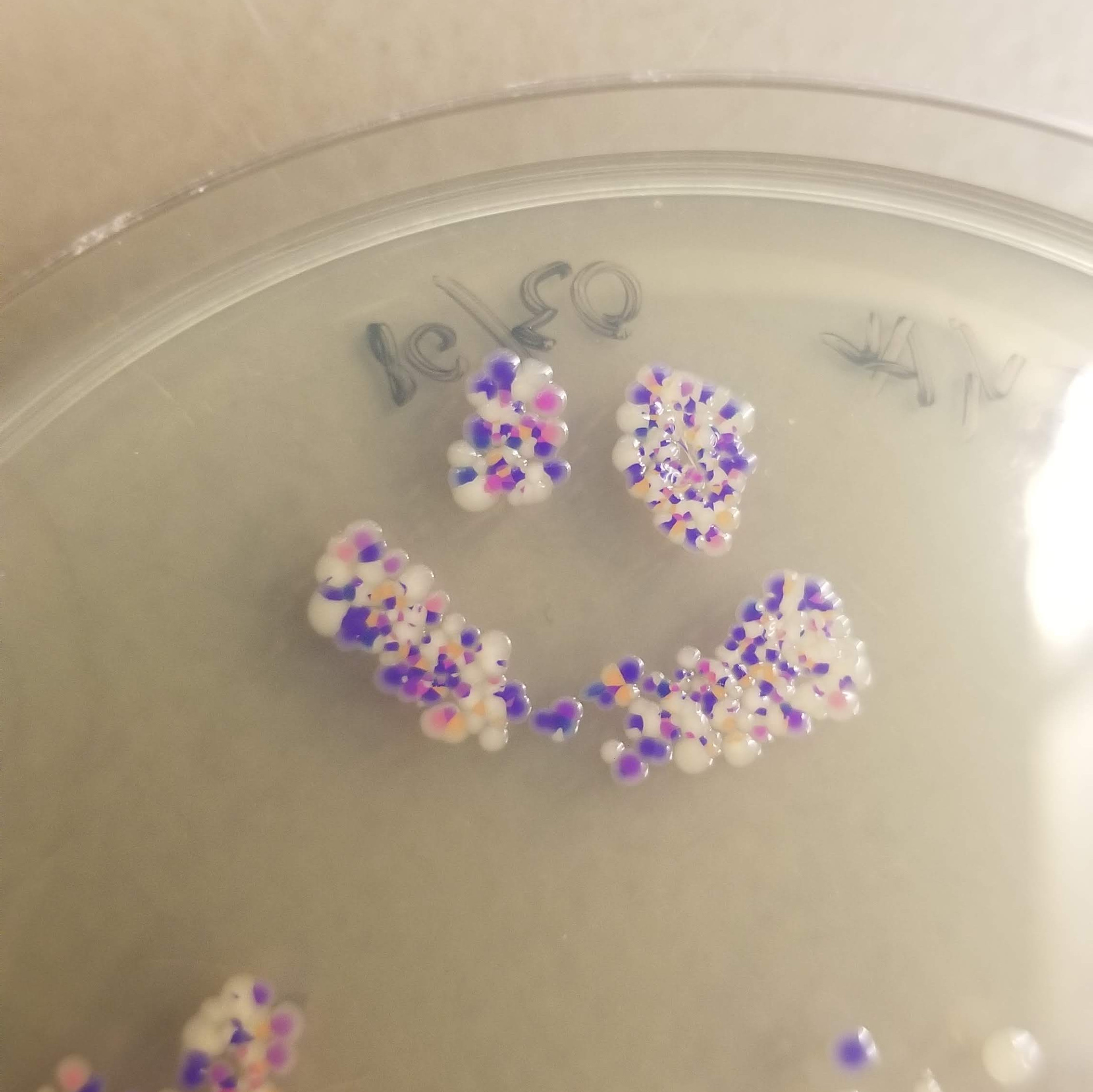
And the results….